Protein-coding gene in the species Homo sapiens
| TAF9 |
|---|
 |
| Identifiers |
|---|
| Aliases | TAF9, MGC:5067, STAF31/32, TAF2G, TAFII-31, TAFII-32, TAFII31, TAFII32, TAFIID32, TATA-box binding protein associated factor 9 |
|---|
| External IDs | OMIM: 600822; MGI: 1888697; HomoloGene: 39986; GeneCards: TAF9; OMA:TAF9 - orthologs |
|---|
| Gene location (Human) |
|---|
 | | Chr. | Chromosome 5 (human)[1] |
|---|
| | Band | 5q13.2 | Start | 69,362,026 bp[1] |
|---|
| End | 69,370,013 bp[1] |
|---|
|
| Gene location (Mouse) |
|---|
 | | Chr. | Chromosome 13 (mouse)[2] |
|---|
| | Band | 13|13 D1 | Start | 100,788,087 bp[2] |
|---|
| End | 100,792,568 bp[2] |
|---|
|
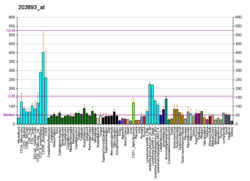
| RNA expression pattern |
|---|
| Bgee | | Human | Mouse (ortholog) |
|---|
| Top expressed in | - left testis
- right testis
- gonad
- islet of Langerhans
- rectum
- superior frontal gyrus
- prefrontal cortex
- monocyte
- dorsolateral prefrontal cortex
- olfactory zone of nasal mucosa
|
| | Top expressed in | - spermatid
- spermatocyte
- testicle
- epiblast
- genital tubercle
- tail of embryo
- secondary oocyte
- zygote
- primary oocyte
- mesencephalon
|
| | More reference expression data |
|
|---|
| BioGPS | 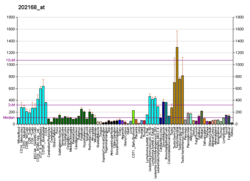
 | | More reference expression data |
|
|---|
|
| Gene ontology |
|---|
| Molecular function | - DNA binding
- ATPase binding
- transcription coactivator activity
- p53 binding
- C2H2 zinc finger domain binding
- histone acetyltransferase activity
- protein binding
- protein heterodimerization activity
- transcription factor binding
- RNA polymerase II general transcription initiation factor activity
| | Cellular component | - transcription factor TFTC complex
- pre-snoRNP complex
- transcription factor TFIID complex
- MLL1 complex
- SAGA complex
- nucleus
- nucleoplasm
| | Biological process | - response to interleukin-1
- regulation of transcription, DNA-templated
- negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator
- protein stabilization
- negative regulation of apoptotic process
- transcription by RNA polymerase II
- transcription, DNA-templated
- cellular response to DNA damage stimulus
- positive regulation of response to cytokine stimulus
- box C/D snoRNP assembly
- positive regulation of cell growth
- histone H3 acetylation
- DNA-templated transcription, initiation
- negative regulation of proteasomal ubiquitin-dependent protein catabolic process
- transcription initiation from RNA polymerase II promoter
- positive regulation of transcription by RNA polymerase II
- snRNA transcription by RNA polymerase II
- regulation of signal transduction by p53 class mediator
- RNA polymerase II preinitiation complex assembly
| | Sources:Amigo / QuickGO |
|
| Orthologs |
|---|
| Species | Human | Mouse |
|---|
| Entrez | | |
|---|
| Ensembl | |
|---|
ENSG00000273841
ENSG00000276463 |
| |
|---|
| UniProt | | |
|---|
| RefSeq (mRNA) | | |
|---|
| RefSeq (protein) | | |
|---|
| Location (UCSC) | Chr 5: 69.36 – 69.37 Mb | Chr 13: 100.79 – 100.79 Mb |
|---|
| PubMed search | [3] | [4] |
|---|
|
| Wikidata |
| View/Edit Human | View/Edit Mouse |
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa, also known as TAF9, is a protein that in humans is encoded by the TAF9 gene.[5][6]
Function
Initiation of transcription by RNA polymerase II requires the activities of more than 70 polypeptides. The protein complex that coordinates these activities is transcription factor IID (TFIID), which binds to the core promoter to position the polymerase properly, serves as the scaffold for assembly of the remainder of the transcription complex, and acts as a channel for regulatory signals. TFIID is composed of the TATA-binding protein (TBP) and a group of evolutionarily conserved proteins known as TBP-associated factors or TAFs. TAFs may participate in basal transcription, serve as coactivators, function in promoter recognition or modify general transcription factors (GTFs) to facilitate complex assembly and transcription initiation. This gene encodes one of the smaller subunits of TFIID that binds to the basal transcription factor GTF2B as well as to several transcriptional activators such as p53 and VP16. A similar but distinct gene (TAF9B) has been found on the X chromosome and a pseudogene has been identified on chromosome 19. Alternative splicing results in multiple transcript variants encoding different isoforms.[5]
Structure
The 17-amino-acid-long trans-activating domains (TAD) of several transcription factors were reported to bind directly to TAF9: p53, VP16, HSF1, NF-IL6, NFAT1, NF-κB, and ALL1/MLL1.[7] Inside of these 17 amino acids, a unique Nine-amino-acid transactivation domain (9aaTAD) was identified for each reported transcription factor.[8] 9aaTAD is a novel domain common to a large superfamily of eukaryotic transcription factors represented by Gal4, Oaf1, Leu3, Rtg3, Pho4, Gln4, Gcn4 in yeast and by p53, NFAT, NF-κB and VP16 in mammals.[9] TAF9 is supposed to be a universal transactivation cofactor for 9aaTAD transcription factors.[8]
Interactions
TAF9 has been shown to interact with:
- GCN5L2,[10]
- Myc,[11]
- SF3B3,[10]
- SUPT7L,[10]
- TADA3L,[10]
- TAF5,[10][12]
- TAF6L,[10]
- TAF10,[10]
- TAF12,[10]
- TAF5L,[10]
- TATA binding protein,[10][13]
- Transcription initiation protein SPT3 homolog,[10] and
- Transformation/transcription domain-associated protein.[10]
References
- ^ a b c ENSG00000276463 GRCh38: Ensembl release 89: ENSG00000273841, ENSG00000276463 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000052293 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b "Entrez Gene: TAF9 TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa".
- ^ Evans SC, Foster CJ, El-Naggar AK, Lozano G (April 1999). "Mapping and mutational analysis of the human TAF2G gene encoding a p53 cofactor". Genomics. 57 (1): 182–3. doi:10.1006/geno.1999.5745. PMID 10191103.
- ^ Uesugi M, Nyanguile O, Lu H, Levine AJ, Verdine GL (August 1997). "Induced alpha helix in the VP16 activation domain upon binding to a human TAF". Science. 277 (5330): 1310–3. doi:10.1126/science.277.5330.1310. PMID 9271577.Uesugi M, Verdine GL (December 1999). "The alpha-helical FXXPhiPhi motif in p53: TAF interaction and discrimination by MDM2". Proc. Natl. Acad. Sci. U.S.A. 96 (26): 14801–6. Bibcode:1999PNAS...9614801U. doi:10.1073/pnas.96.26.14801. PMC 24728. PMID 10611293.Choi Y, Asada S, Uesugi M (May 2000). "Divergent hTAFII31-binding motifs hidden in activation domains". J. Biol. Chem. 275 (21): 15912–6. doi:10.1074/jbc.275.21.15912. PMID 10821850.Venot C, Maratrat M, Sierra V, Conseiller E, Debussche L (April 1999). "Definition of a p53 transactivation function-deficient mutant and characterization of two independent p53 transactivation subdomains". Oncogene. 18 (14): 2405–10. doi:10.1038/sj.onc.1202539. PMID 10327062.Lin J, Chen J, Elenbaas B, Levine AJ (May 1994). "Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein". Genes Dev. 8 (10): 1235–46. doi:10.1101/gad.8.10.1235. PMID 7926727.
- ^ a b Piskacek S, Gregor M, Nemethova M, Grabner M, Kovarik P, Piskacek M (June 2007). "Nine-amino-acid transactivation domain: establishment and prediction utilities". Genomics. 89 (6): 756–68. doi:10.1016/j.ygeno.2007.02.003. PMID 17467953.
- ^ The prediction for 9aa TADs (for both acidic and hydrophilic transactivation domains) is available online from National EMBnet-Node Austria ("9aaTAD Prediction Webtool". EMBnet AUSTRIA. Archived from the original on 2007-07-01. Retrieved 2009-01-10.)
- ^ a b c d e f g h i j k l Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG (Oct 2001). "Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo". Mol. Cell. Biol. 21 (20): 6782–95. doi:10.1128/MCB.21.20.6782-6795.2001. PMC 99856. PMID 11564863.
- ^ Liu X, Tesfai J, Evrard YA, Dent SY, Martinez E (May 2003). "c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation". J. Biol. Chem. 278 (22): 20405–12. doi:10.1074/jbc.M211795200. PMC 4031917. PMID 12660246.
- ^ Tao Y, Guermah M, Martinez E, Oelgeschläger T, Hasegawa S, Takada R, Yamamoto T, Horikoshi M, Roeder RG (Mar 1997). "Specific interactions and potential functions of human TAFII100". J. Biol. Chem. 272 (10): 6714–21. doi:10.1074/jbc.272.10.6714. PMID 9045704.
- ^ Bellorini M, Lee DK, Dantonel JC, Zemzoumi K, Roeder RG, Tora L, Mantovani R (Jun 1997). "CCAAT binding NF-Y-TBP interactions: NF-YB and NF-YC require short domains adjacent to their histone fold motifs for association with TBP basic residues". Nucleic Acids Res. 25 (11): 2174–81. doi:10.1093/nar/25.11.2174. PMC 146709. PMID 9153318.
Further reading
- Klemm RD, Goodrich JA, Zhou S, Tjian R (1995). "Molecular cloning and expression of the 32-kDa subunit of human TFIID reveals interactions with VP16 and TFIIB that mediate transcriptional activation". Proc. Natl. Acad. Sci. U.S.A. 92 (13): 5788–92. Bibcode:1995PNAS...92.5788K. doi:10.1073/pnas.92.13.5788. PMC 41586. PMID 7597030.
- Hisatake K, Ohta T, Takada R, Guermah M, Horikoshi M, Nakatani Y, Roeder RG (1995). "Evolutionary conservation of human TATA-binding-polypeptide-associated factors TAFII31 and TAFII80 and interactions of TAFII80 with other TAFs and with general transcription factors". Proc. Natl. Acad. Sci. U.S.A. 92 (18): 8195–9. Bibcode:1995PNAS...92.8195H. doi:10.1073/pnas.92.18.8195. PMC 41123. PMID 7667268.
- Lu H, Levine AJ (1995). "Human TAFII31 protein is a transcriptional coactivator of the p53 protein". Proc. Natl. Acad. Sci. U.S.A. 92 (11): 5154–8. Bibcode:1995PNAS...92.5154L. doi:10.1073/pnas.92.11.5154. PMC 41867. PMID 7761466.
- Thut CJ, Chen JL, Klemm R, Tjian R (1995). "p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60". Science. 267 (5194): 100–4. doi:10.1126/science.7809597. PMID 7809597.
- Zhou Q, Sharp PA (1995). "Novel mechanism and factor for regulation by HIV-1 Tat". EMBO J. 14 (2): 321–8. doi:10.1002/j.1460-2075.1995.tb07006.x. PMC 398086. PMID 7835343.
- Parada CA, Yoon JB, Roeder RG (1995). "A novel LBP-1-mediated restriction of HIV-1 transcription at the level of elongation in vitro". J. Biol. Chem. 270 (5): 2274–83. doi:10.1074/jbc.270.5.2274. PMID 7836461.
- Ou SH, Garcia-Martínez LF, Paulssen EJ, Gaynor RB (1994). "Role of flanking E box motifs in human immunodeficiency virus type 1 TATA element function". J. Virol. 68 (11): 7188–99. doi:10.1128/JVI.68.11.7188-7199.1994. PMC 237158. PMID 7933101.
- Kashanchi F, Piras G, Radonovich MF, Duvall JF, Fattaey A, Chiang CM, Roeder RG, Brady JN (1994). "Direct interaction of human TFIID with the HIV-1 transactivator tat". Nature. 367 (6460): 295–9. Bibcode:1994Natur.367..295K. doi:10.1038/367295a0. PMID 8121496. S2CID 4362048.
- Adams MD, Soares MB, Kerlavage AR, Fields C, Venter JC (1993). "Rapid cDNA sequencing (expressed sequence tags) from a directionally cloned human infant brain cDNA library". Nat. Genet. 4 (4): 373–80. doi:10.1038/ng0893-373. PMID 8401585. S2CID 12612300.
- Wang Z, Morris GF, Rice AP, Xiong W, Morris CB (1996). "Wild-type and transactivation-defective mutants of human immunodeficiency virus type 1 Tat protein bind human TATA-binding protein in vitro". J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 12 (2): 128–38. doi:10.1097/00042560-199606010-00005. PMID 8680883.
- Pendergrast PS, Morrison D, Tansey WP, Hernandez N (1996). "Mutations in the carboxy-terminal domain of TBP affect the synthesis of human immunodeficiency virus type 1 full-length and short transcripts similarly". J. Virol. 70 (8): 5025–34. doi:10.1128/JVI.70.8.5025-5034.1996. PMC 190456. PMID 8764009.
- Kashanchi F, Khleif SN, Duvall JF, Sadaie MR, Radonovich MF, Cho M, Martin MA, Chen SY, Weinmann R, Brady JN (1996). "Interaction of human immunodeficiency virus type 1 Tat with a unique site of TFIID inhibits negative cofactor Dr1 and stabilizes the TFIID-TFIIA complex". J. Virol. 70 (8): 5503–10. doi:10.1128/JVI.70.8.5503-5510.1996. PMC 190508. PMID 8764062.
- Zhou Q, Sharp PA (1996). "Tat-SF1: cofactor for stimulation of transcriptional elongation by HIV-1 Tat". Science. 274 (5287): 605–10. Bibcode:1996Sci...274..605Z. doi:10.1126/science.274.5287.605. PMID 8849451. S2CID 13266489.
- Tao Y, Guermah M, Martinez E, Oelgeschläger T, Hasegawa S, Takada R, Yamamoto T, Horikoshi M, Roeder RG (1997). "Specific interactions and potential functions of human TAFII100". J. Biol. Chem. 272 (10): 6714–21. doi:10.1074/jbc.272.10.6714. PMID 9045704.
- García-Martínez LF, Ivanov D, Gaynor RB (1997). "Association of Tat with purified HIV-1 and HIV-2 transcription preinitiation complexes". J. Biol. Chem. 272 (11): 6951–8. doi:10.1074/jbc.272.11.6951. PMID 9054383.
- Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, Howard BH, Qin J, Nakatani Y (1998). "Histone-like TAFs within the PCAF histone acetylase complex". Cell. 94 (1): 35–44. doi:10.1016/S0092-8674(00)81219-2. PMID 9674425. S2CID 18942972.
- Vassilev A, Yamauchi J, Kotani T, Prives C, Avantaggiati ML, Qin J, Nakatani Y (1999). "The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily". Mol. Cell. 2 (6): 869–75. doi:10.1016/S1097-2765(00)80301-9. PMID 9885574.
- Evans SC, Foster CJ, El-Naggar AK, Lozano G (1999). "Mapping and mutational analysis of the human TAF2G gene encoding a p53 cofactor". Genomics. 57 (1): 182–3. doi:10.1006/geno.1999.5745. PMID 10191103.
- Lai CH, Chou CY, Ch'ang LY, Liu CS, Lin W (2000). "Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics". Genome Res. 10 (5): 703–13. doi:10.1101/gr.10.5.703. PMC 310876. PMID 10810093.
- Choi Y, Asada S, Uesugi M (2000). "Divergent hTAFII31-binding motifs hidden in activation domains". J. Biol. Chem. 275 (21): 15912–6. doi:10.1074/jbc.275.21.15912. PMID 10821850.

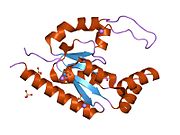 1rkb: The structure of adrenal gland protein AD-004
1rkb: The structure of adrenal gland protein AD-004


















